Research
Decoding Immune function at the atomic level
Decoding Immune function at the atomic level
At Sgourakis Lab, we study systems which play a fundamental role in adaptive immune responses. Our group’s principal efforts focus on mechanistic, structural and functional studies of protein complexes, with emphasis on the development of an iterative experimental / computational approach.
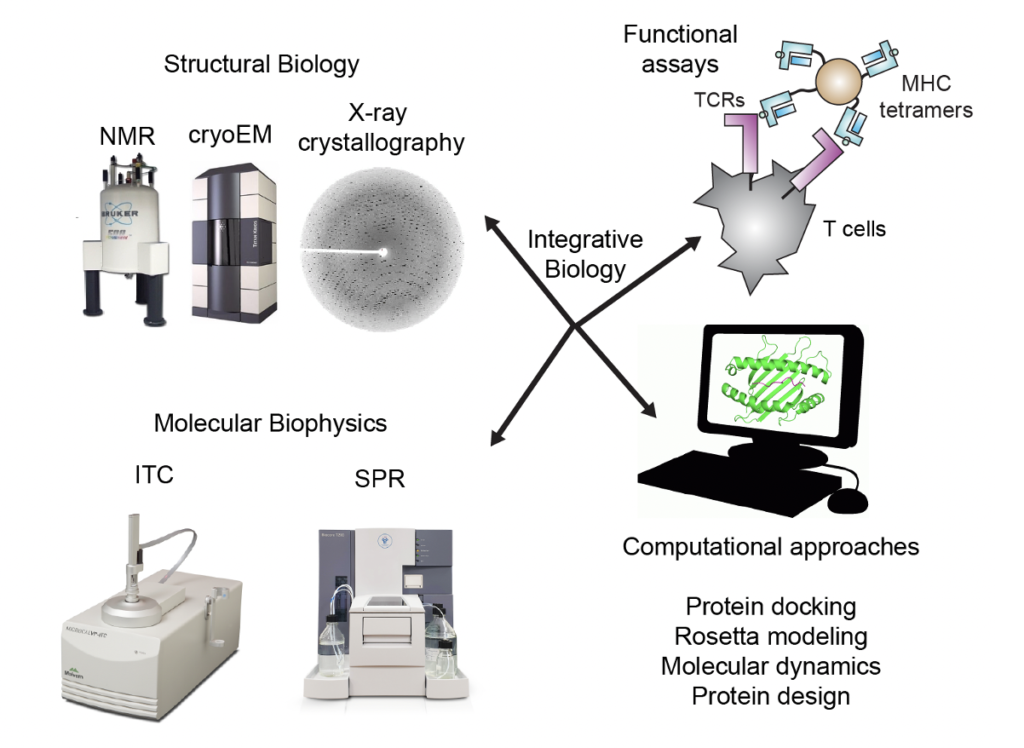
Sgourakis Lab’s research is directed at characterizing and manipulating molecular recognition of protein complexes of the innate and adaptive immune systems. Principal focuses in the lab include antigen processing and presentation by the Major Histocompatibility Complex (MHC), the role of immune chaperones in the assembly of the MHC and effects on antigen repertoires, recognition of antigen/MHC complexes by T cell receptors (TCRs), pleiotropic cytokines function and dynamics, development of new tools to characterize T cell repertoires in diseased patients, and engineering of novel immunotherapies based on T cell-mediated immunity. Because the MHC encodes the most polymorphic protein in the human genome (>3,000 allotypes identified in humans), and is associated with more diseases than any other gene, our research has important translational potential. Unravelling how antigens are loaded onto the MHC and subsequently recognized by immune chaperones and TCR receptors will help us understand autoimmune diseases, such as diabetes, multiple sclerosis and arthritis, and immune responses to invading pathogens and developing tumors. Moreover, obtaining atomic resolution insights into complex interactions, characterizing biomolecular dynamics, and determining pathways for conformational exchange not only reveals insights into biological function, but provides molecular blueprints to develop new therapeutics. Our integrative approach melds together structural biology, molecular biophysics, biochemistry, protein design, computational biology, and cancer immunology.
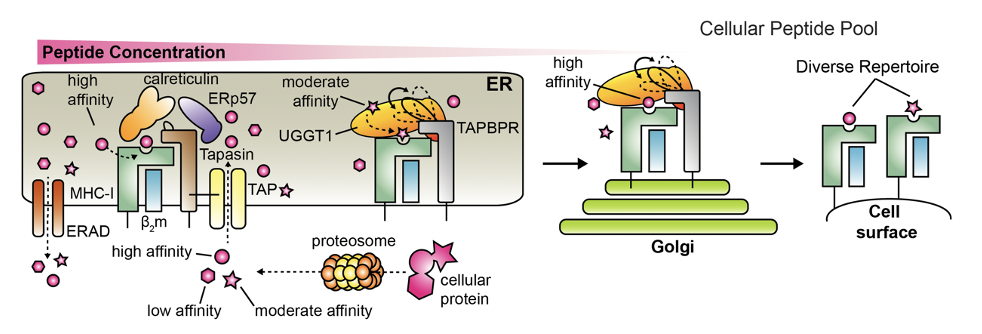
Our lab has spent the last several years examining a plethora of aspects for chaperone/MHC interactions, mostly with respect to the most recently identified immune chaperone, TAPBPR. First, we have described the role of switch in the MHC (called the alpha2-1 helix), which undergoes significant conformational changes during a “tug-of-war” between the incoming peptide antigen and the TAPBPR chaperone. Interestingly, for the peptide antigen to win the “tug-of-war”, allosteric communication between MHC sites spanning the antigen binding groove are required (McShan et al, Nature Chemical Biology, 2018). This study provided the first high-resolution picture of the peptide/MHC/TAPBPR intermediate complex, and provided insights into the molecular rules that define the selection of high-affinity peptides.
Second, we have examined how amino acid polymorphisms in the MHC, called HLA in humans, dictate chaperone recognition and antigen selection. In collaboration with the Procko lab (University of Illinois), We performed deep mutagenesis studies in human cell lines, and experimentally determined protein dynamics in chaperone-dependent and chaperone-independent HLA proteins and correlated observed conformational changes with chaperone recognition (McShan, Devlin et al, PNAS, 2019). This study showed that protein dynamics are molecular determinants to the significant differences in how different HLA proteins are recognized by chaperones, and provides insights into how antigen presentation and chaperone recognition influence disease susceptibility between patients with different HLA proteins.
Third, we have characterized the role of a loop in TAPBPR and revealed its unique function as a peptide trap to lower the free energy of peptide binding (McShan et al, Nature Communications, 2021). This study provided a new mechanism into how chaperones can help shape antigen repertoires.

Human leukocyte antigens (HLAs) are essential peptide-presenting complexes relevant to immune response and dysregulated in several human diseases, including cancer. While HLAs are among the most polymorphic genes in humans, several HLA:peptide complexes have been identified as potential neoantigens in human diseases and represent promising therapeutic targets. In our HLA research, we investigate the potential of peptide-centric biologics to selectively recognize and target HLA:peptide complexes present in human diseases. While these peptide-centric biologics are difficult to design without cross-reactivity to other endogenous HLA complexes, the Maris lab at CHOP has spearheaded the development of a chimeric antigen receptor (CAR) which binds an HLA:peptide target expressed exclusively in neuroblastoma. Through X-ray crystallography and computational modeling, we’ve identified that the identified peptide residues are solvently exposed and accessible for CAR recognition and association (Yarmarkovich et al., Nature 2021). We now seek to utilize a combination of X-ray crystallography, molecular dynamics simulations, and computational modeling to characterize the structural basis of specificity for additionally identified peptide-centric biologics, which may lead to targeted therapies for human diseases. This work is carried out in close collaboration with the group of pediatric oncologist Dr. John Maris and postdoctoral scientist Dr. Mark Yarmarkovich.
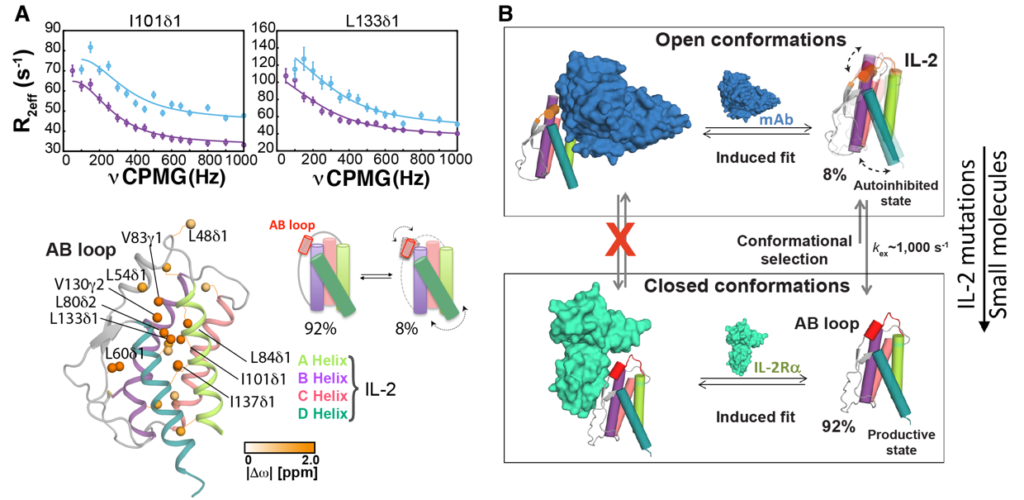
Interleukin-2 is a multifunctional signaling molecule relevant to physiological and dysregulated disease states including autoimmunity and cancer, however efforts to exploit it for therapeutic purposes have been hampered by serious side effects. In our IL-2 research, we explore – on an atomic level – the rare conformational states that IL-2 transiently adopts as it functions. These states are difficult to capture and visualize but our laboratory employed state-of-the-art NMR spectroscopy techniques to show that IL-2 adopts two different structural forms that affect how it interacts with the receptors on different types of T cells. In solution, IL-2 naturally shifts back and forth between a minor conformation and a major conformation (De Paula et al., PNAS 2020). We are now extending this approach to the human IL-2 system with the goal to obtain a high-resolution view of the IL-2 conformational landscape. In addition, our laboratory aims to develop IL-2 mimetics which recapitulate the conformational features of the Treg-biasing state, and ultimately lead to more targeted therapies for autoimmune disease treatment.